Okay… Back to take a stab at this
I have not come up with a good “science-free” analogy so we are going to have to get a little in the weeds here, please bear with me
First, and most importantly, I AM NOT REFERENCING A BALL PYTHON GENOME HERE!! I had to find something that roughly applied and that I had a passing familiarity with so that I knew I was not talking out my own backside. I am not claiming that this is the absolute answer for these ball genes in question. This is just a possible explanation using another organism as reference
With that out of the way…
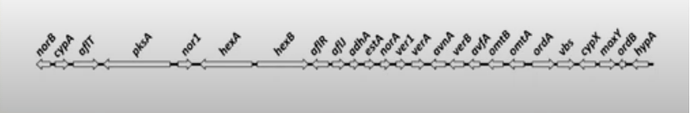
This graphic represents a cluster of genes known as an operon
Operons are clusters of related genes that are all involved in a single process. In this specific operon, every one of these genes is necessary for the production of a single end product
Because all of the genes play an important role, they tend to be regulated as a complete unit. Frequently, that regulation happens at the role of transcription (copying DNA into RNA). One of the ways this is accomplished is having a single start point for transcription that creates a single RNA containing all the genes in the operon instead of making multiple smaller transcripts. Think of it kind of like copying a whole chapter out of a book in one go as opposed to copying each individual paragraph, one at a time, on different sheets of paper
If we look at the graphic again, let us say the identified mutations all fall in the gene labeled vbs. Without vbs, the critical product of the operon is never made and so we see a mutant phenotype
So… What about the outliers?
What happens if I throw a mutation in the small space between norB and cypA? Conventional “herp hobby wisdom” is that it does not matter because those genes are completely unrelated to vbs
But…
Remember what I said above about operons being transcribed as a single uint?
What if the mutation between norB and cypA results in the transcription process stopping there? In that case, vbs (along with a bunch of other things) will not get made, and so we see a mutant phenotype again
And if I have an animal that carries one chromosome with the mutation to vbs and another chromosome with a mutation between norB and cypA, what will the offspring from that animal come out as?
Half of them will inherit the vbs mutation and the other half will inherit the norB and cypA spacer mutation.
This outcome would basically mirror what you would see if you had an animal carrying two different mutant copies of vbs, right?
So… This then would be an example of how mutations can be non-allelic while still behaving in an allelic manner
Does that make sense??